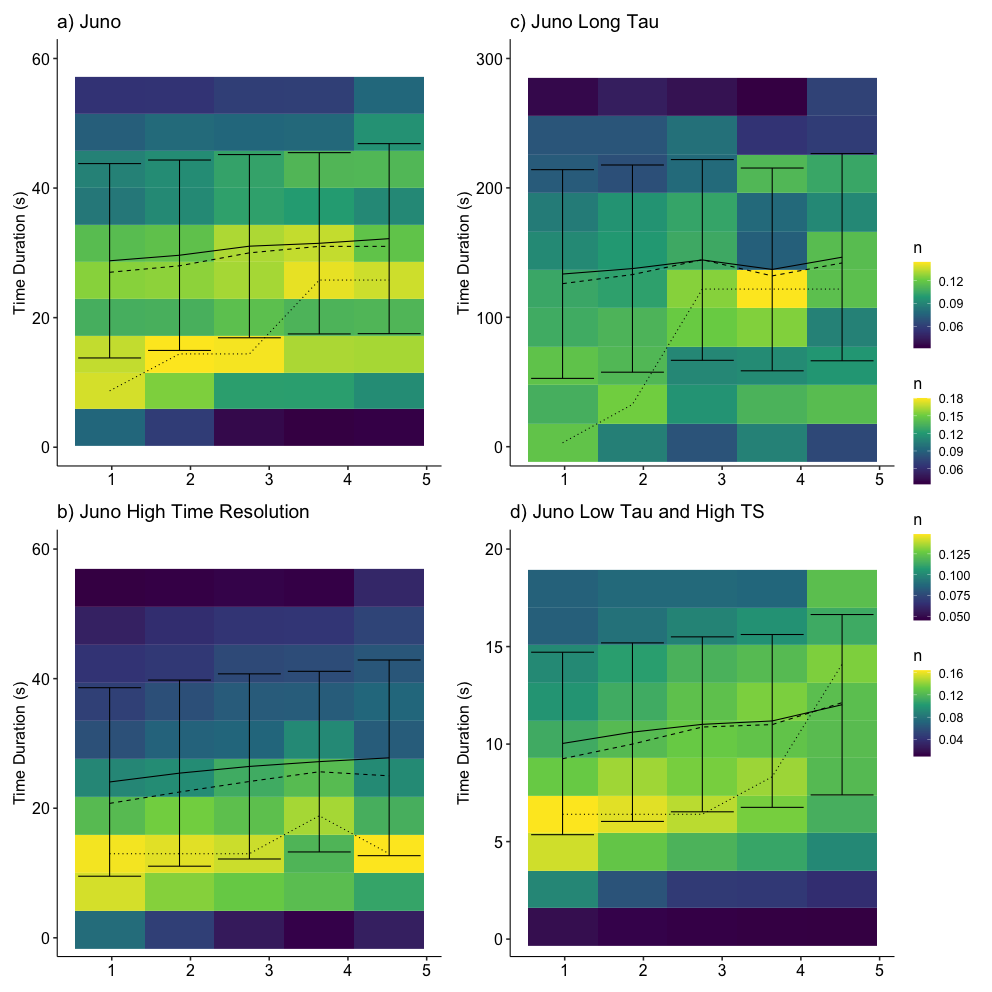
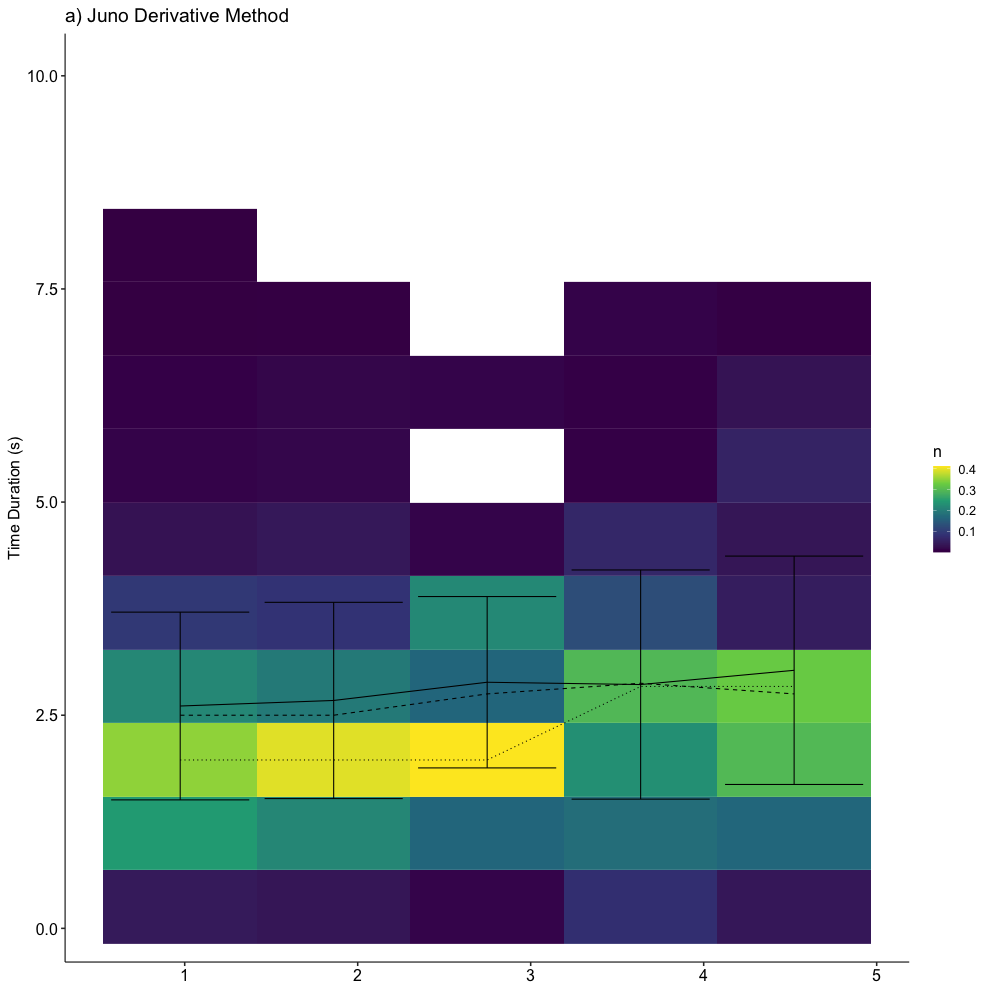
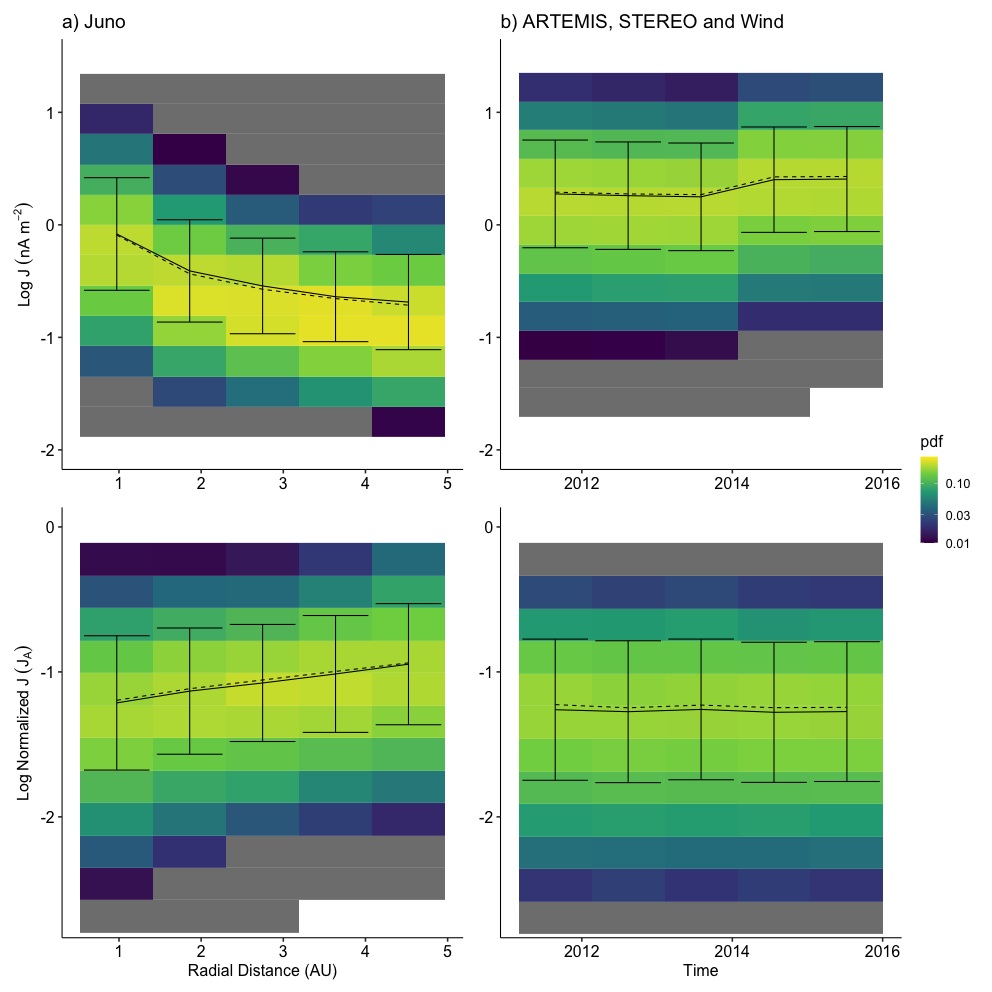
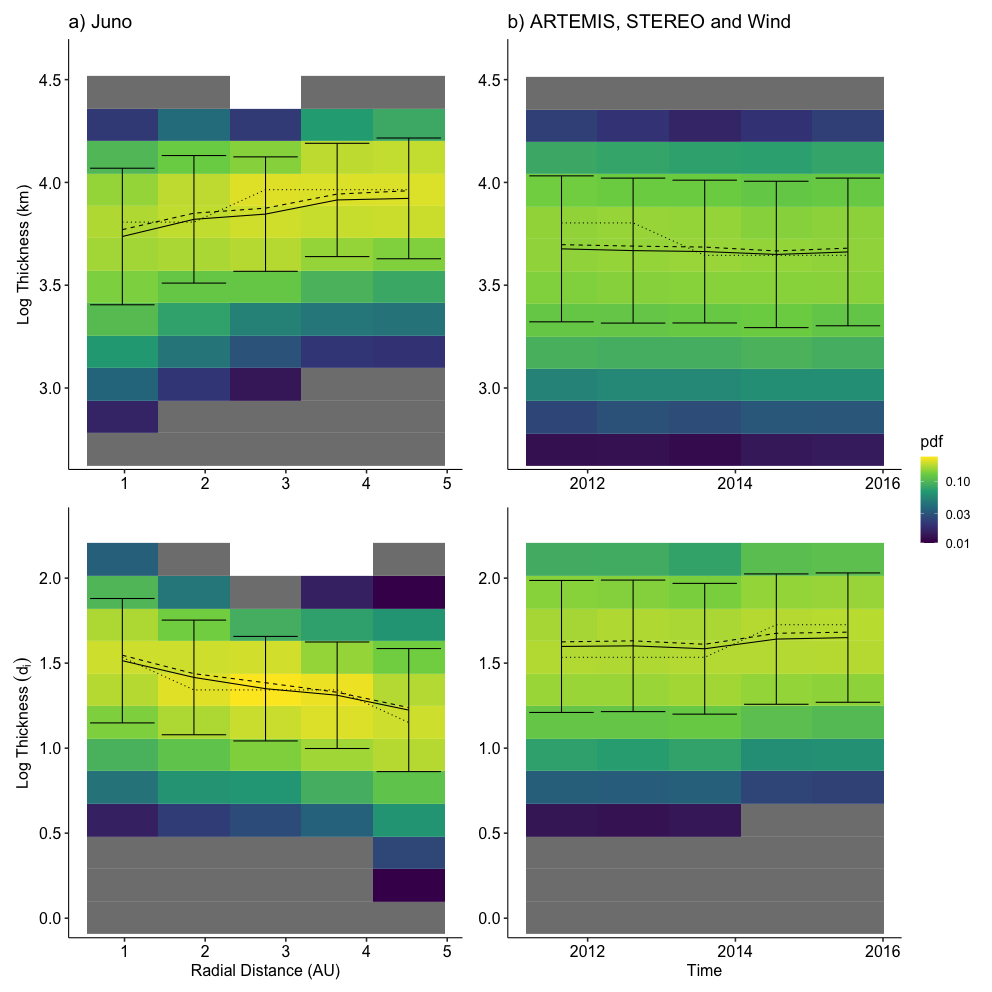
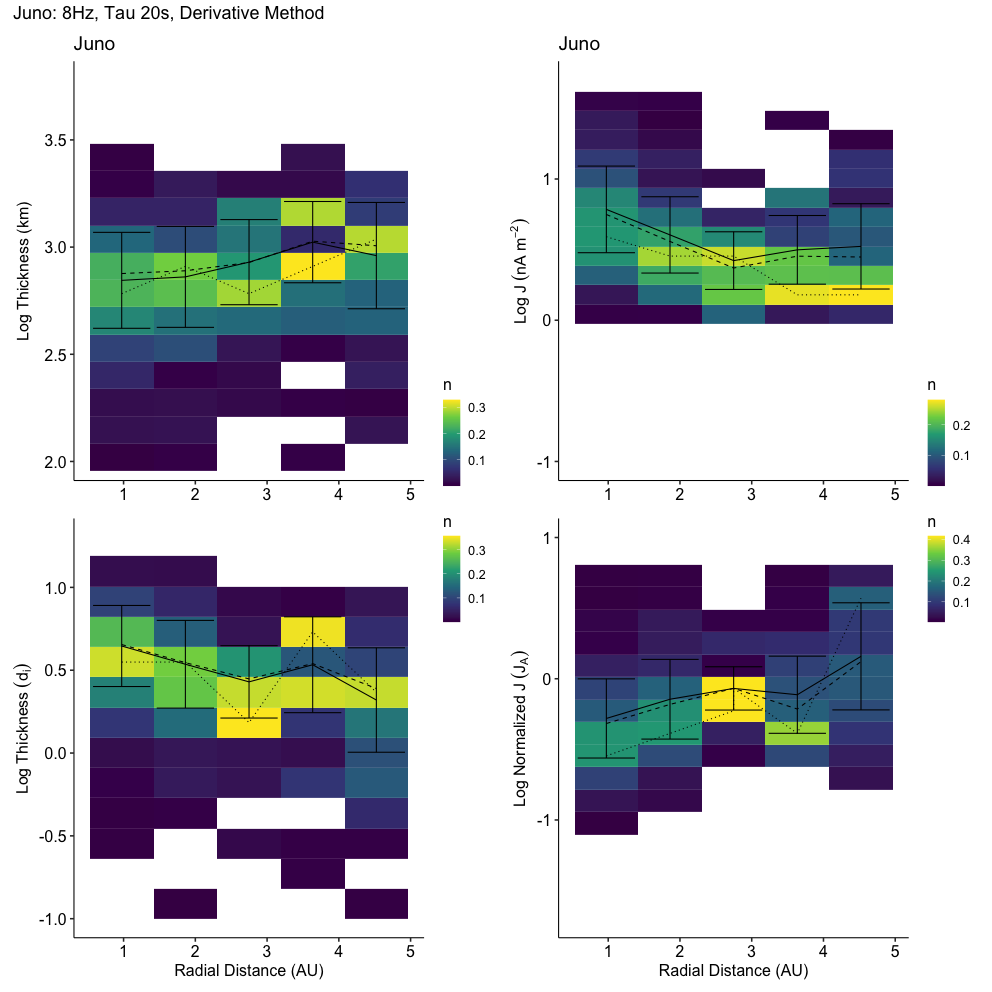
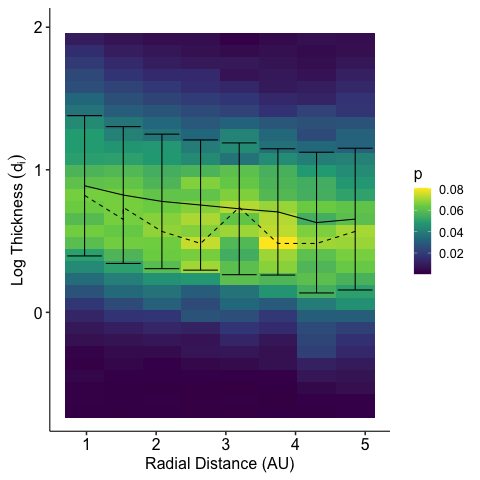
2024-02-18 21:55:57.510 | INFO | __main__:<module>:13 - Columns: ['time', 'tstart', 'tstop', 'd_tstart', 'd_tstop', 'd_time', 'time_before', 'time_after', 'count', 'B_std', 'B_mean', 'dB_vec', 'index_diff', 'index_std', 'index_fluctuation', 'B.after', 'B.before', 'b_mag', 'b_n', 'bn_over_b', 'd_star', 'db_mag', 'db_over_b', 'db_over_b_max', 'fit.stat.chisqr', 'fit.stat.rsquared', 'fit.vars.amplitude', 'fit.vars.c', 'fit.vars.sigma', 'rotation_angle', 'dB_x', 'dB_y', 'dB_z', 'dB_lmn_x', 'dB_lmn_y', 'dB_lmn_z', 'k_x', 'k_y', 'k_z', 'Vl_x', 'Vl_y', 'Vl_z', 'Vn_x', 'Vn_y', 'Vn_z', 'duration', 'radial_distance', 'plasma_density', 'plasma_temperature', 'model_b_r', 'model_b_t', 'model_b_n', 'v_x', 'v_y', 'v_z', 'plasma_speed', 'B_background_x', 'B_background_y', 'B_background_z', 'v_x_before', 'v_y_before', 'v_z_before', 'n.before', 'v.ion.before', 'plasma_temperature_before', 'v_x_after', 'v_y_after', 'v_z_after', 'n.after', 'v.ion.after', 'plasma_temperature_after', 'v_l', 'v_n', 'v_k', 'v_mn', 'L_k', 'j0_k', 'ion_inertial_length', 'Alfven_speed', 'j_Alfven', 'L_k_norm', 'j0_k_norm', 'v.ion.before.l', 'v.ion.after.l', 'B.vec.before.l', 'B.vec.before.m', 'B.vec.before.n', 'B.vec.after.l', 'B.vec.after.m', 'B.vec.after.n', 'v.Alfven.before', 'v.Alfven.after', 'v.Alfven.before.l', 'v.Alfven.after.l', 'n.change', 'v.ion.change', 'v.ion.change.l', 'B.change', 'v.Alfven.change', 'v.Alfven.change.l', 'sat', 'ts', 'method', 'ts_method', 'label', 'index_d_diff']