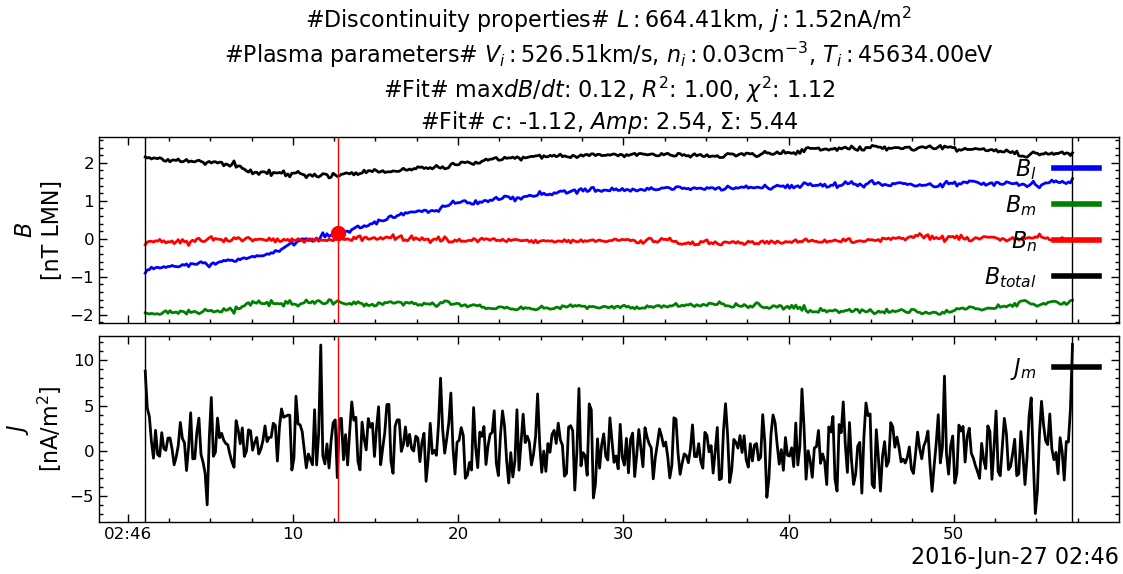
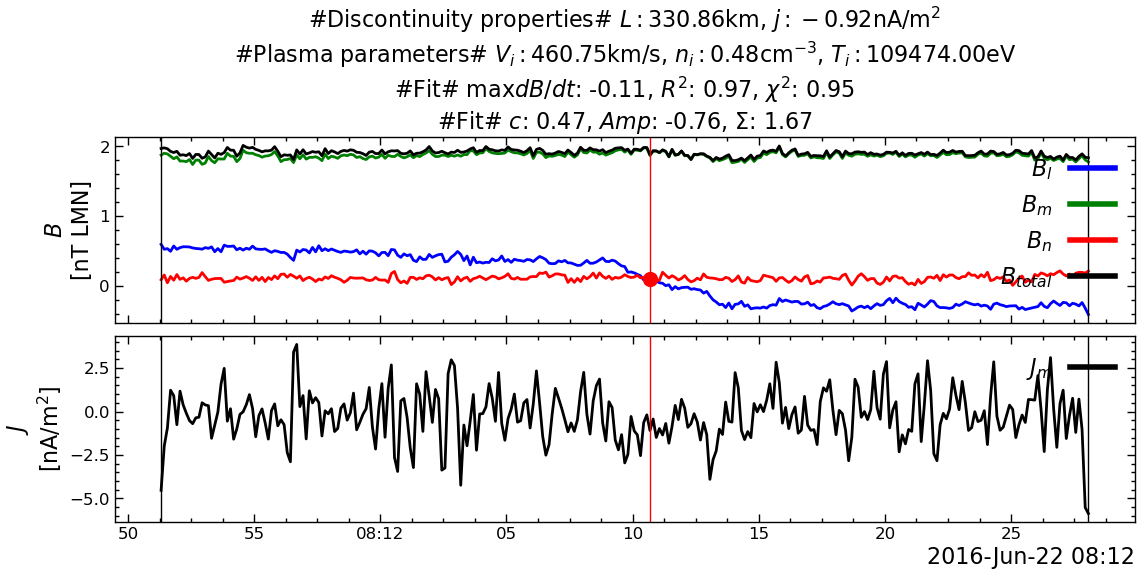
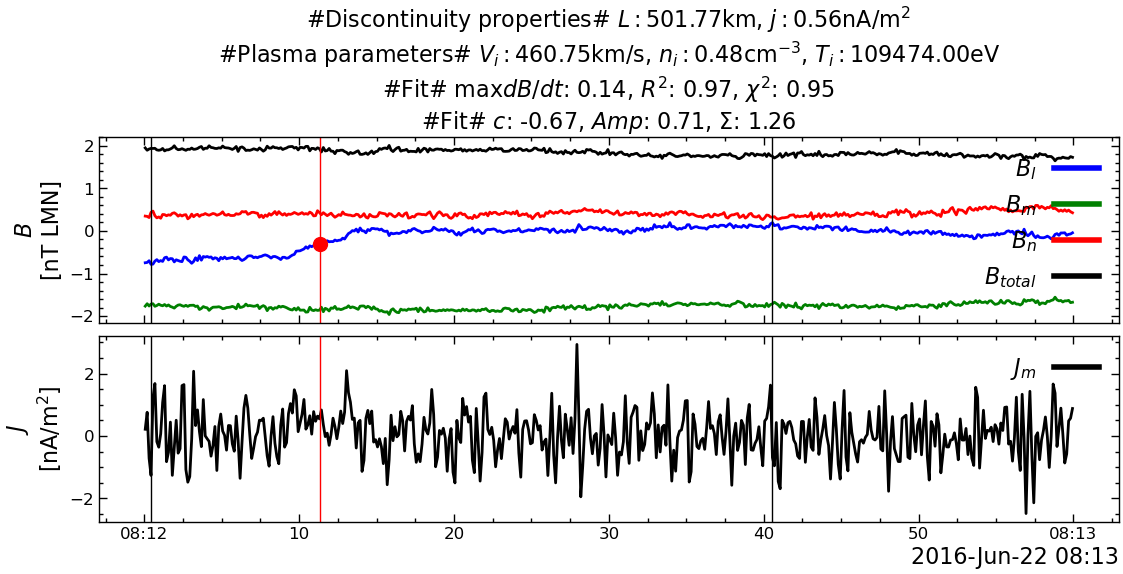
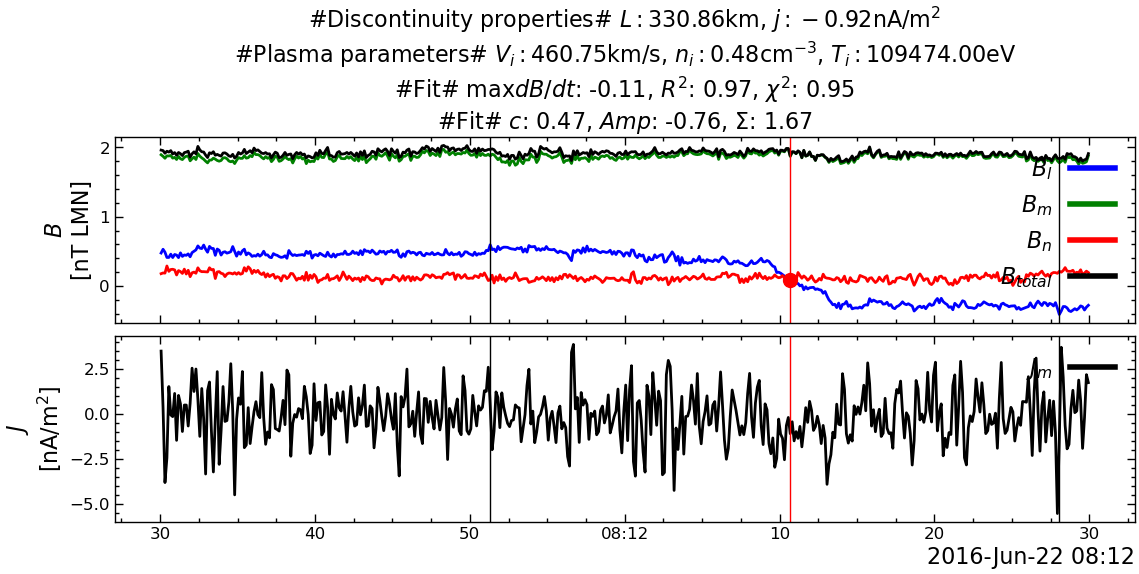
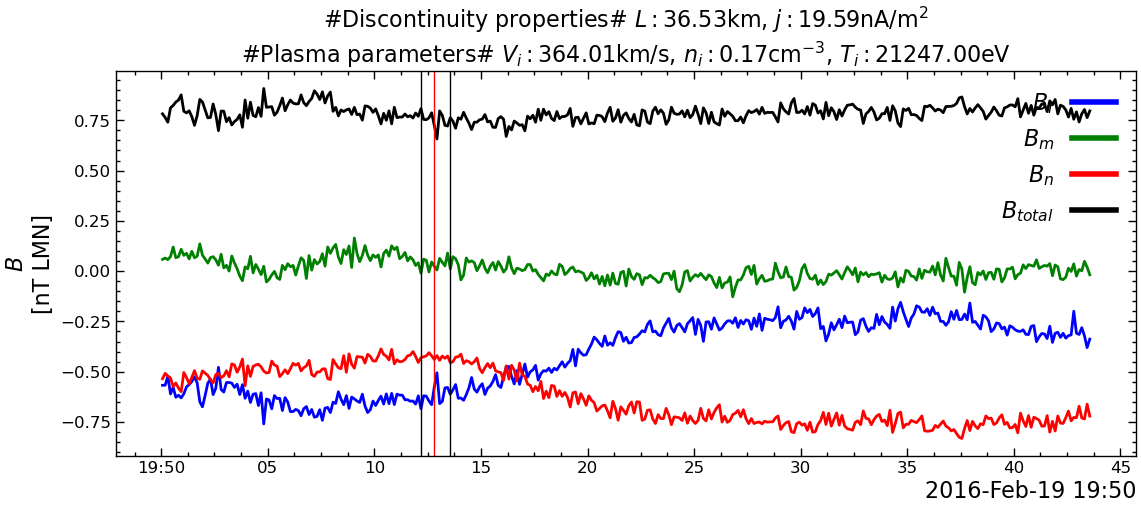
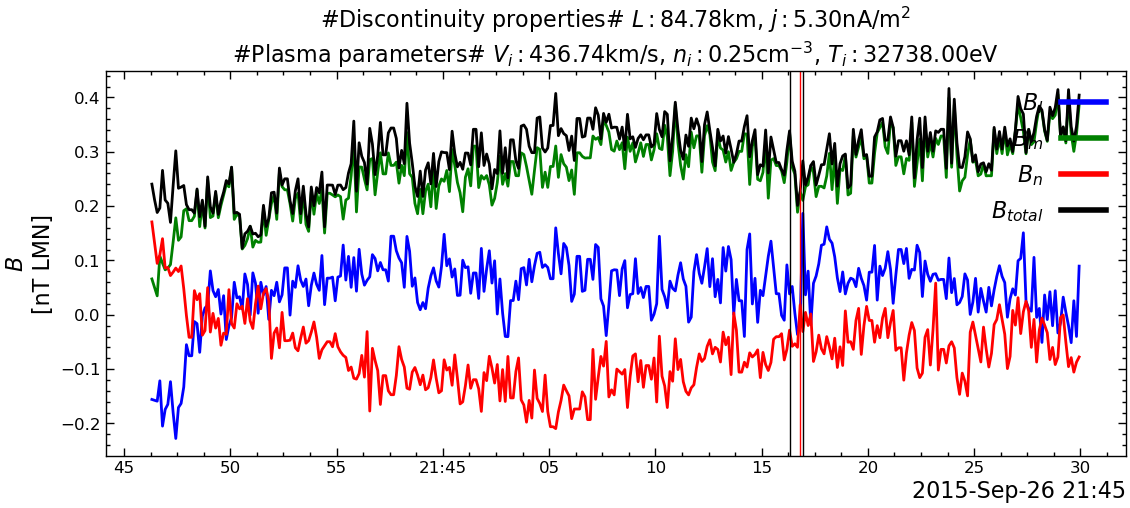
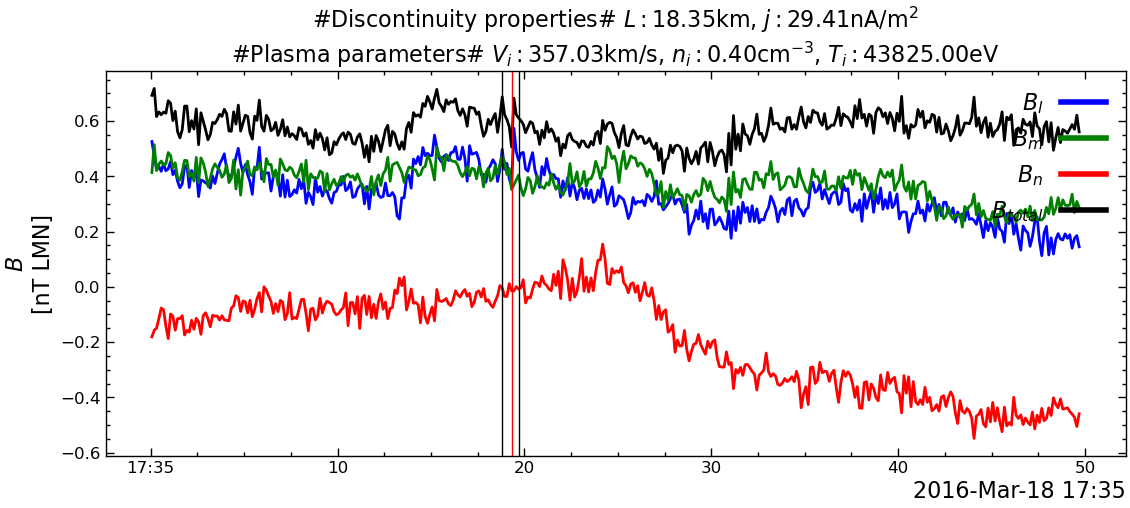
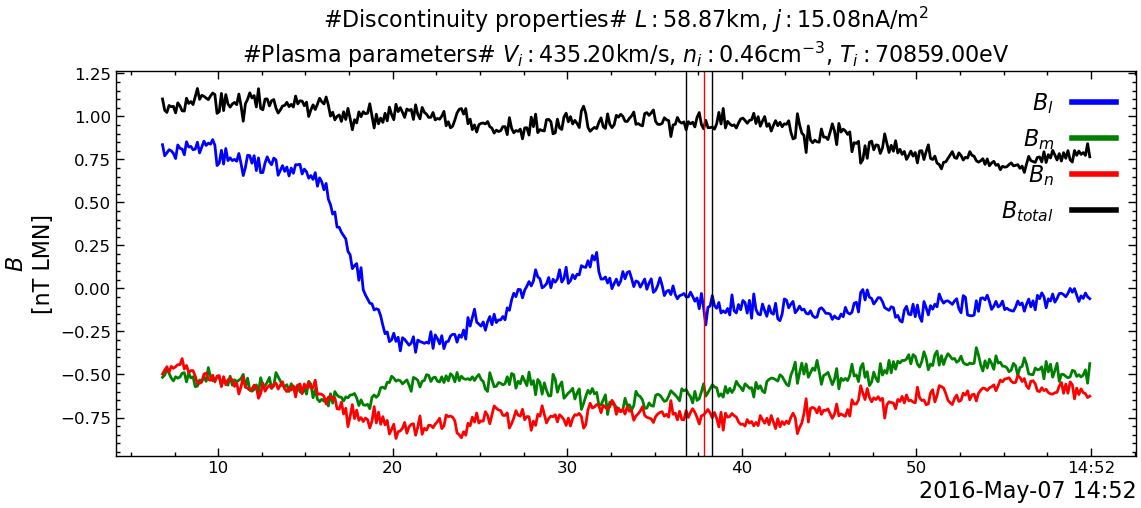
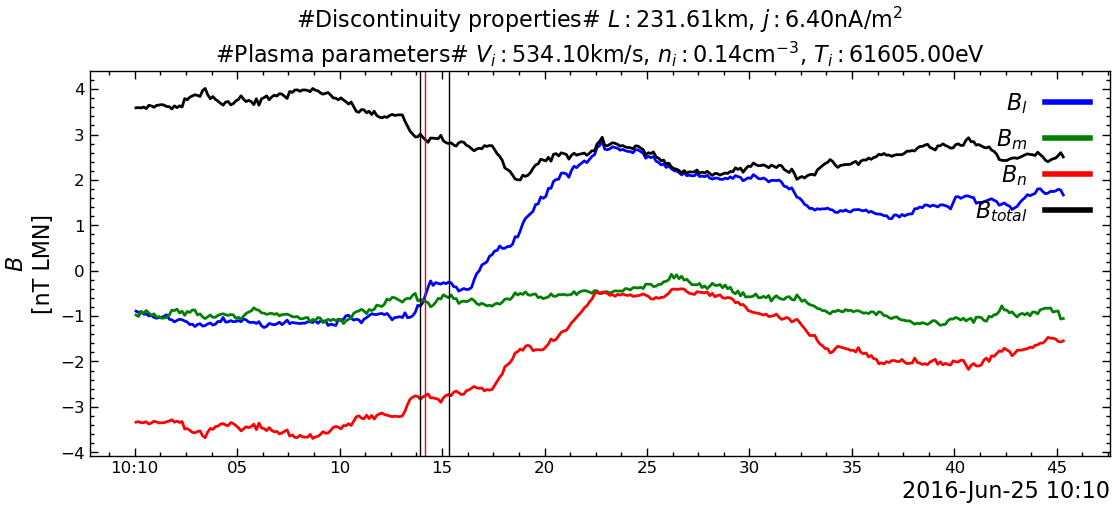
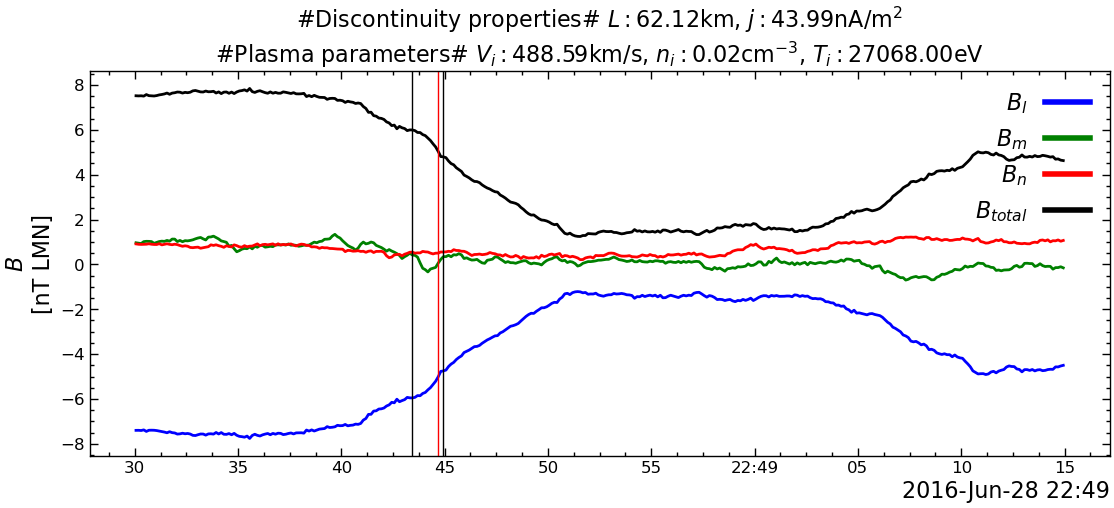
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[35], line 4
1 #| layout-ncol: 3
2 #| column: screen
3 offset = timedelta(seconds=30)
----> 4 figs = j_config.plot_candidates(num=15, add_plasma_params=True, offset=offset)
File ~/micromamba/envs/psp_conjunction/lib/python3.11/site-packages/discontinuitypy/datasets.py:245, in IDsDataset.plot_candidates(self, indices, num, random, predicate, **kwargs)
242 indices = indices.head(num).to_numpy()
243 logger.info(f"Candidates indices: {indices}")
--> 245 return [self.plot_candidate(index=i, **kwargs) for i in indices]
File ~/micromamba/envs/psp_conjunction/lib/python3.11/site-packages/discontinuitypy/datasets.py:245, in <listcomp>(.0)
242 indices = indices.head(num).to_numpy()
243 logger.info(f"Candidates indices: {indices}")
--> 245 return [self.plot_candidate(index=i, **kwargs) for i in indices]
File ~/micromamba/envs/psp_conjunction/lib/python3.11/site-packages/discontinuitypy/datasets.py:224, in IDsDataset.plot_candidate(self, event, index, **kwargs)
222 if event is None:
223 event = self.get_event(index)
--> 224 data = self.get_event_data(event, **kwargs)
226 return _plot_candidate(event, data, **kwargs)
File ~/micromamba/envs/psp_conjunction/lib/python3.11/site-packages/discontinuitypy/datasets.py:100, in IdsEvents.get_event_data(self, event, offset, **kwargs)
97 end = event["t.d_end"] + offset
99 _data = self.data.filter(pl.col("time").is_between(start, end))
--> 100 return df2ts(_data, self.bcols)
File ~/micromamba/envs/psp_conjunction/lib/python3.11/site-packages/discontinuitypy/utils/basic.py:292, in df2ts(df, cols, time_col, attrs, name)
289 cols = df.columns.tolist()
291 if isinstance(df, pl.LazyFrame):
--> 292 df = df.collect()
294 # Prepare data
295 data = df[cols].to_numpy()
File ~/micromamba/envs/psp_conjunction/lib/python3.11/site-packages/polars/lazyframe/frame.py:1940, in LazyFrame.collect(self, type_coercion, predicate_pushdown, projection_pushdown, simplify_expression, slice_pushdown, comm_subplan_elim, comm_subexpr_elim, no_optimization, streaming, background, _eager)
1937 if background:
1938 return InProcessQuery(ldf.collect_concurrently())
-> 1940 return wrap_df(ldf.collect())
KeyboardInterrupt: