begin
using DrWatson
using FHist
using HDF5
using StatsBase: sample
using DataFrames
using DimensionalData
using DimensionalData: dims
using StatsBase: mean
using Beforerr
import CairoMakie.Makie.SpecApi as S
include("../src/io.jl")
include("../src/plot.jl")
include("../src/jump.jl")
include("../src/mix_rates.jl")
TM_OBS_FILE = "tm_obs.jld2"
VMIN = 0.25
VMAX = 256
endIn [0]:
Mixing rates
Transition matrix approach
In [0]:
using EasyFit
# Load the data
d = wload(datadir(TM_OBS_FILE))
df = d["df"]
vPs = d["vPs"]
tms = d["tms"]
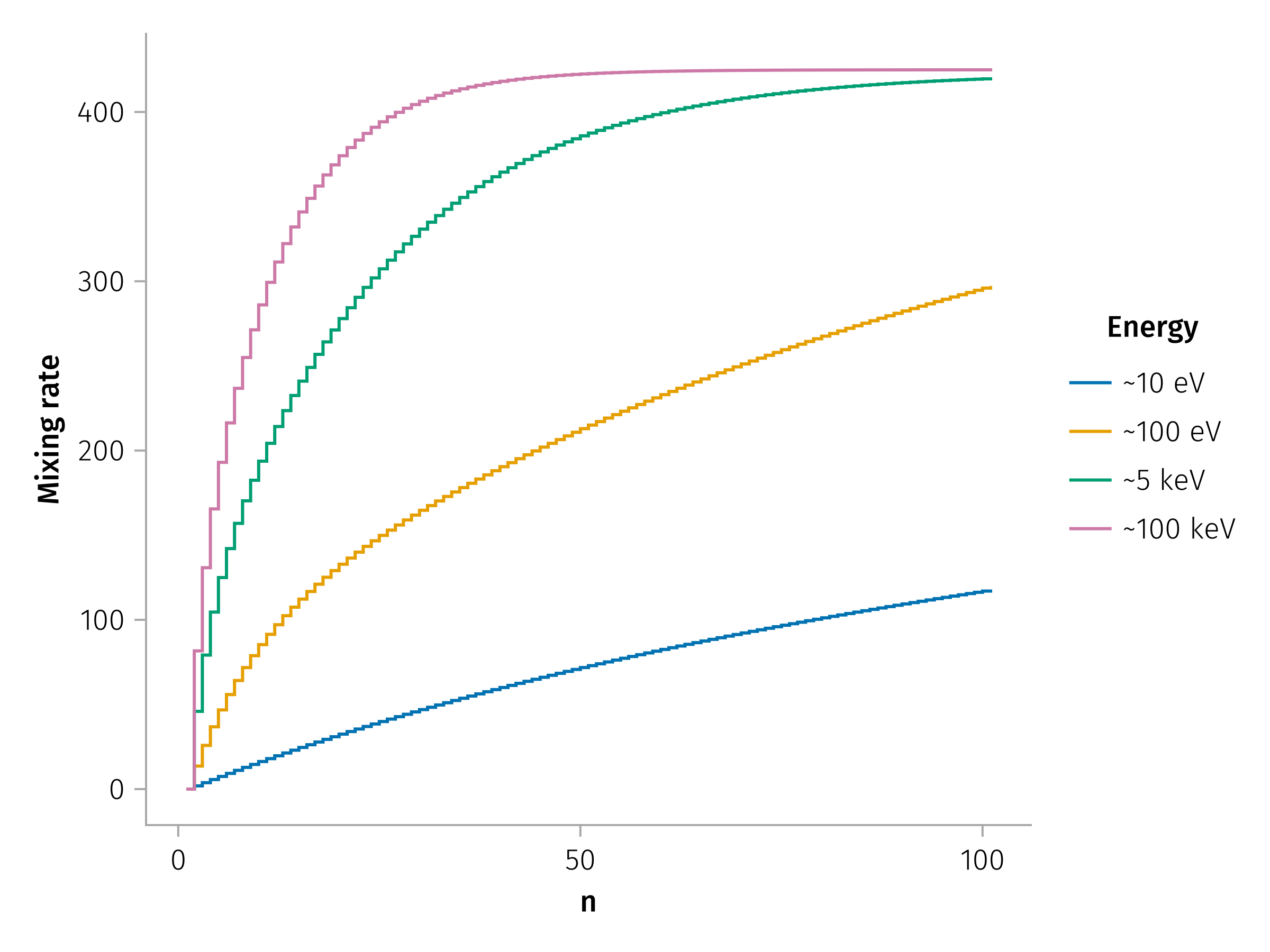
ergs_approx = ["~10 eV", "~100 eV", "~5 keV", "~100 keV", "~1 MeV"]
r = 0:200
res = map(tms) do tm
mix_rate(tm).(r)
end
fit_label(f) = L"D_{μμ} = %$(round(1 / f.b, digits=2))"
xlabel = L"n"
# Plot the mixing rate
function plot_mixing_rate!(res)
options = Options(nbest=8, besttol=1e-5)
map(res) do re
N = length(re)
sca = scatter!(1:N, re)
fit = fitexp(1:N, re; options)
lin = lines!(fit.x, fit.y)
[sca, lin, fit]
end
end
f1 = let labels = ergs_approx, ax = (; xlabel, ylabel="M_2(∞) - M_2(n)", xscale=log10, yscale=log10)
data = map(res[2:end]) do re
last(re) .- re
end
f = Figure()
ax = Axis(f[1, 1]; ax...)
contents = plot_mixing_rate!(data)
labels = LaTeXString.(ergs_approx[2:end] .* "(" .* fit_label.(getindex.(contents, 3)) .* ")")
contents = getindex.(contents, Ref([1, 2]))
axislegend(ax, contents, labels, "Energy"; position=:rb)
xlims!(1.5, 200)
ylims!(3e-2, 3e-1)
end
f2 = let labels = ergs_approx, ax = (; xlabel, ylabel=L"M_2(n)", xscale=log10, yscale=log10)
data = res[2:end]
f = Figure()
ax = Axis(f[1, 1]; ax...)
contents = plot_mixing_rate!(data)
labels = LaTeXString.(ergs_approx[2:end] .* "(" .* fit_label.(getindex.(contents, 3)) .* ")")
contents = getindex.(contents, Ref([1, 2]))
axislegend(ax, contents, labels, "Energy"; position=:rb)
xlims!(1.5, 200)
ylims!(3e-2, 3e-1)
easy_save("mixing_rate")
end
f2ECDF approach (better)
In [0]:
dir = "simulations"
dfs = get_result_dfs(; dir)In [0]:
foreach(get_result!, eachrow(dfs))
@rtransform!(dfs, :ecdf_da = ecdf_da(:results, "s2α0", "Δs2α"))
@rtransform!(dfs, :ecdf_μ = ecdf_da(:results, "μ0", "Δμ"))In [0]:
function load_obs_hist(; file=datadir("obs/") * "wind_hist3d.h5")
h = h5readhist(file, "hist")
bc = bincenters(h)
vs = 2 .^ bc[1]
βs = bc[2] ./ 2
θs = bc[3]
return DimArray(bincounts(h), (v=vs, β=βs, θ=θs))
end
function dargmax(p)
# Note: iteration is deliberately unsupported for CartesianIndex.
name.(dims(p)), map(getindex, dims(p), Tuple(argmax(p)))
end
function check_obs_hist(p)
# check the maxximum value of the observation data
@info maximum(p)
@info dargmax(p)
end
p = load_obs_hist()
function sample_hist(p, args...)
a = []
wv = Float64[]
foreach(Iterators.product(dims(p)...)) do t
push!(a, t)
push!(wv, p[v=At(t[1]), β=At(t[2]), θ=At(t[3])])
end
sample(a, Weights(wv), args...)
end
In [0]:
function subset_ecdf(v0, β, θ, vP; df=dfs, col=:ecdf_da, vmin=VMIN, vmax=VMAX)
v = clamp(vP / v0, vmin, vmax)
sdf = @subset(df, :v .== v, :β .== β, :θ .== θ; view=true)
ecdf_das = sdf[!, col]
if isempty(ecdf_das)
@warn "No data found for v=$v, β=$β, θ=$θ, v0=$v0"
elseif length(ecdf_das) != 1
@warn "Multiple data found for v=$v, β=$β, θ=$θ, v0=$v0"
end
return ecdf_das[1]
end
function rand_jumps(x0, vP, p; n=10, kw...)
x = [x0]
samples = sample_hist(p, n)
for i in 1:n
ecdf_da = subset_ecdf(samples[i]..., vP; kw...)
x_new = x[end] + rand_jump(x[end], ecdf_da)
push!(x, x_new)
end
return x
endIn [0]:
vPs = 4 .^ (3:6)
counts = 1024
n = 100
vPs_jumps = map(vPs) do vP
xs = range(0, 1, length=counts)
jumps = rand_jumps.(xs, vP, Ref(p); n)
(; vP, jumps)
end
vPs_jumps_ani = map(vPs) do vP
xs = range(0.4, 0.6, length=counts)
jumps = rand_jumps.(xs, vP, Ref(p); n)
(; vP, jumps)
end
vPs_μJumps = map(vPs) do vP
xs = range(-1, 1, length=counts)
jumps = rand_jumps.(xs, vP, Ref(p); n, col=:ecdf_μ)
(; vP, jumps)
endIn [0]:
function jumps_ecdfplot(jumps; idxs=[1, div(size(jumps, 1), 2), size(jumps, 1)], kw...)
linestyles = [:solid, :dash, :dot]
ps = map(enumerate(idxs)) do (i, idx)
S.ECDFPlot(jumps[idx, :]; linestyle=linestyles[i], kw...)
end
S.Axis(; plots=ps, xlabel="sin(α)^2", ylabel="F")
end
function jumps_ecdfplot_grid(jumps_vs; kw...)
ax1 = jumps_ecdfplot(stack(jumps_vs[1].jumps); color=Cycled(1), kw...)
ax2 = jumps_ecdfplot(stack(jumps_vs[2].jumps); color=Cycled(2), kw...)
ax3 = jumps_ecdfplot(stack(jumps_vs[3].jumps); color=Cycled(3), kw...)
ax4 = jumps_ecdfplot(stack(jumps_vs[4].jumps); color=Cycled(4), kw...)
S.GridLayout([ax1 ax2; ax3 ax4])
end
In [0]:
f = Figure(;)
ax = Axis(f[1, 1]; xlabel="n", ylabel="M2")
contents = map(vPs_jumps) do t
vP, jumps = t
m2s = m2(jumps)
scatterlines!(m2s; label="vP=$vP")
end
labels = ergs_approx[1:4]
ax = Axis(f[2, 1]; xlabel="n", ylabel="M2")
contents = map(vPs_jumps_ani) do t
vP, jumps = t
m2s = m2(jumps)
scatterlines!(m2s; label="vP=$vP")
end
idxs = [1, 2, 4]
gl1 = jumps_ecdfplot_grid(vPs_jumps; idxs)
gl2 = jumps_ecdfplot_grid(vPs_jumps_ani; idxs)
gl = S.GridLayout([gl1, gl2])
plot(f[1:end, 2], gl)
Legend(f[1:end, 3], contents, labels, "Energy")
easy_save("mixing_rate_sin2")
# let jumps = s2α_jumps, n = 8
# sample_jumps = sample(jumps, n)
# scatterlines!(vec(m2))
# ax = Axis(f[2, 1]; xlabel = "n", ylabel = "sin(α)^2")
# stairs!.(sample_jumps)
# f
# endIn [0]:
D_nn(m2s) = m2s[2] - m2s[1]
vPs_m2s = map(vPs_μJumps) do t
vP, jumps = t
m2(jumps)
end
D_nns = D_nn.(vPs_m2s)
result_df = DataFrame(vP=vPs, m2s=vPs_m2s, D_nn=D_nns)In [0]:
labels = ergs_approx[1:4]
contents = map(vPs_jumps_ani) do t
vP, jumps = t
m2s = m2(jumps)
S.ScatterLines(m2s; label="vP=$vP")
end
gl1 = S.GridLayout([S.Axis(; plots=contents, xlabel="n", ylabel="M2")])
gl2 = jumps_ecdfplot_grid(vPs_jumps_ani; idxs)
plot(S.GridLayout([gl1, gl2], rowsizes=[Auto(2), Auto(3)]))
easy_save("mr/mixing_rate_sin2_ani")